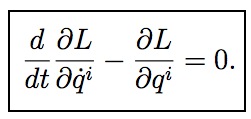I’ve mentioned their book the “Structure and Interpretation of Classical Mechanics” a couple times already; today I’d like to recommend that you read a wonderful short paper by Gerald Jay Sussman and Jack Wisdom, called “The Role of Programming in the Formulation of Ideas,” which helps explain why understanding physics and the other mathematical sciences can sometimes be so difficult. The basic point is that our notation is often an absolute mess, caused by the fact that we use equations like we use natural language, in a highly ambiguous way:
“It is necessary to present the science in the language of mathematics. Unfortunately, when we teach science we use the language of mathematics in the same way that we use our natural language. We depend upon a vast amount of shared knowledge and culture, and we only sketch an idea using mathematical idioms.”
The solution proposed is to develop notation that can be understood by computers, which do not tolerate ambiguity:
“One way to become aware of the precision required to unambiguously communicate a mathematical idea is to program it for a computer. Rather than using canned programs purely as an aid to visualization or numerical computation, we use computer programming in a functional style to encourage clear thinking. Programming forces one to be precise and formal, without being excessively rigorous. The computer does not tolerate vague descriptions or incomplete constructions. Thus the act of programming makes one keenly aware of one’s errors of reasoning or unsupported conclusions.”
Sussman and Wisdom then focus on one highly illuminating example, the Lagrange equations. These equations can be derived from the fundamental principle of least action. This principle tells you that if you have a classical system that begins in a configuration C1 at time t1 and arrives at a configuration C2 at time t2, the path it traces out between t1 and t2 will be the one that is consistent with the initial and final configurations and minimizes the integral over time of the Lagrangian for the system, where the Lagrangian is given by the kinetic energy minus the potential energy.
Physics textbooks tell us that if we apply the calculus of variations to the integral of the Lagrangian (called the “action”) we can derive that the true path satisfies the Lagrange equations, which are traditionally written as:
Here L is the Lagrangian, t is the time, annd qi are the coordinates of the system.
These equations (and many others like them) have confused and bewildered generations of physics students. What is the problem? Well, there are all sorts of fundamental problems in interpreting these equations, detailed in Sussman and Wisdom’s paper. As they point out, basic assumptions like whether a coordinate and its derivative are independent variables are not consistent within the same equation. And shouldn’t this equation refer to the path somewhere, since the Lagrange equations are only correct for the true path? I’ll let you read Sussman and Wisdom’s full laundry list of problems yourself. But let’s turn to the psychological effects of using such equations:
“Though such statements (and derivations that depend upon them) seem very strange to students, they are told that if they think about them harder they will understand. So the student must either come to the conclusion that he/she is dumb and just accepts it, or that the derivation is correct, with some appropriate internal rationalization. Students often learn to carry out these manipulations without really understanding what they are doing.”
Is this true? I believe it certainly is (my wife agrees: she gave up on mathematics, even though she always received excellent grades, because she never felt she truly understood). The students who learn to successfully rationalize such ambiguous equations, and forget about the equations that they can’t understand at all, are the ones who might go on to be successful physicists. Here’s an example, from the review of Sussman and Wisdom’s book by Piet Hut, a very well-regarded physicist who is now a professor at the Institute for Advanced Studies:
“… I went through the library in search of books on the variational principle in classical mechanics. I found several heavy tomes, borrowed them all, and started on the one that looked most attractive. Alas, it didn’t take long for me to realize that there was quite a bit of hand-waving involved. There was no clear definition of the procedure used for computing path integrals, let alone for the operations of differentiating them in various ways, by using partial derivatives and/or using an ordinary derivative along a particular path. And when and why the end points of the various paths had to be considered fixed or open to variation also was unclear, contributing to the overall confusion.
Working through the canned exercises was not very difficult, and from an instrumental point of view, my book was quite clear, as long as the reader would stick to simple examples. But the ambiguity of the presentation frustrated me, and I started scanning through other, even more detailed books. Alas, nowhere did I find the clarity that I desired, and after a few months I simply gave up. Like generations of students before me, I reluctantly accepted the dictum that ‘you should not try to understand quantum mechanics, since that will lead you astray for doing physics’, and going even further, I also gave up trying to really understand classical mechanics! Psychological defense mechanisms turned my bitter sense of disappointment into a dull sense of disenchantment.”
Sussman and Wisdom do show how the ambiguous conventional notation can be replaced with unambiguous notation that can even be used to program a computer. Because it’s new, it will feel alien at first; the Lagrange equations look like this:
It’s worth learning Sussman and Wisdom’s notation for the clarity it ultimately provides. It’s even more important to learn to always strive for clear understanding.
One final point: although mathematicians do often use notation that is superior to physicists’, they shouldn’t feel too smug; Sussman and Wisdom had similar things to say about differential geometry in this paper.